INTRODUCTION
Several physicochemical procedures are used for the assessment of the quality attributes of Insulin Human and various insulin analogues, hereafter called insulins. Of these procedures, peptide mapping and determination of high molecular weight proteins are similar for the various insulin drug substances and drug products. This chapter describes the tests for peptide mapping and for quantitative analysis of high molecular weight proteins that can be used for the various insulins.
Specific instructions that deviate from the general procedures outlined here are given in the respective drug substance or drug product monographs for the different insulins.
PEPTIDE MAPPING
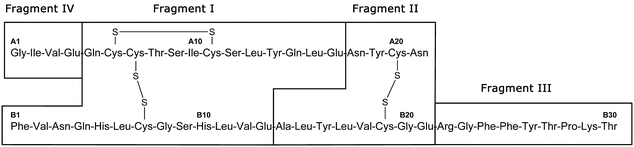
Insulin digestion is performed using Staphylococcus aureus strain V8 (S. aureus V8) protease, a serine endoproteinase that cleaves at the C-terminal side of glutamyl and aspartyl residues in phosphate buffer at pH 7.8. This protease is specific for Glu-C digestion in ammonium bicarbonate or other non-phosphate–containing buffers at pH 7.8. The presence of proline on the carboxy side of the peptide bond inhibits cleavage. The buffer system used should cleave all glutamyl bonds in insulin without cleavage of the aspartyl bonds. In general, insulin is digested into four peptides in the buffer system described in the Sample solution below.
The following show the amino acid differences of the insulin analogues compared to Insulin Human and the in silico differences in the fragments obtained upon digestion with S. aureus V8.
Figure 1 shows the four fragments of Insulin Human following digestion with S. aureus V8 protease.
Table 1 shows the amino acid differences between insulin analogues and Insulin Human.
Table 1. Amino Acid Differences between Insulin Analogues and Insulin Human
| Insulin Analogue
|
Amino Acid Differences from Insulin Human |
|---|---|
| Insulin aspart | B28 Asp |
| Insulin glargine | A21 Gly, two Arg added to C-terminus of B chain |
| Insulin glulisine | B3 Lys, B29 Glu |
| Insulin lispro | B28 Lys, B29 Pro |
Table 2 shows specific S. aureus V8 protease digest fragments of insulin analogues. Amino acid differences compared to Insulin Human are highlighted.
Table 2. Amino Acid Differences: Insulin Human and Analogues (Differences with Insulin Human Are Shown in Boldface)
| Insulin Analogue | Amino Acid Differences Compared to Insulin Humana | |
|---|---|---|
| Insulin aspart | Fragment III | (B22) Arg-Gly-Phe-Phe-Tyr-Thr-Asp-Lys-Thr (B30) |
| Insulin glargine | Fragment II | (A18) Asn-Tyr-Cys-Gly (A21) |
| Fragment III | (B22) Arg-Gly-Phe-Phe-Tyr-Thr-Pro-Lys-Thr-Arg-Arg (B32) | |
| Insulin glulisine | Fragment I | (B1) Phe-Val-Lys-Gln-His-Leu-Cys-Gly-Ser-His-Leu (B11) |
| Fragment III | (B22) Arg-Gly-Phe-Phe-Tyr-Thr-Pro-Glu-Thr (B30) | |
| Insulin lispro | Fragment III | (B22) Arg-Gly-Phe-Phe-Tyr-Thr-Lys-Pro-Thr (B30) |
|
a
A and B denote the A and B chains of insulin, respectively; numbers denote amino acid position in the chain.
|
||
• Peptide Mapping Procedure
The following procedure is applicable for preparing peptide maps of Insulin Human and insulin analogues.
HEPES buffer:
Dissolve 2.38 g of HEPES (N-2-hydroxyethylpiperazine-N¢-2-ethanesulfonic acid) in about 90 mL of water in a 100-mL volumetric flask. Adjust with 5 M sodium hydroxide to a pH of 7.5, dilute with water to volume, and mix.
Sulfate buffer:
2.0 M ammonium sulfate and 0.5 M sulfuric acid (1:1). Mix, and filter.
• Insulin Digestion
The following procedure provides efficient cleavage of the glutamyl bonds of insulin. [Note—Volumes up to 20-fold higher can be used as long as the ratio of the solutions remains the same. If interfering autolysis byproducts are observed in the chromatogram when a digest of the enzyme alone is run, the enzyme:insulin ratio must be decreased and digestion time must be increased. ]
Enzyme solution:
Prepare a 1-mg/mL solution of S. aureus V8 protease in water (approximately 500 units/mg).
Sample solution:
Prepare a 2.0-mg/mL solution of the insulin to be examined in 0.01 N hydrochloric acid. To a clean vial add 25 µL of the 2.0-mg/mL insulin solution, 100 µL of HEPES buffer, and 20 µL of Enzyme solution (final ratio is 25:100:20). Cap the vial, and incubate at 25 for 6 h. Stop the reaction by adding an equal volume of Sulfate buffer. Longer incubation times may be needed for analogues with poor solubility at pH 7.5.
for 6 h. Stop the reaction by adding an equal volume of Sulfate buffer. Longer incubation times may be needed for analogues with poor solubility at pH 7.5.
Standard solution:
Prepare at the same time and in the same manner a solution of the appropriate USP Insulin Reference Standard as directed for the Sample solution.
• Peptide Fragment Determination
Determine the peptide fragments using the following peptide-mapping procedure (see Biotechnology-Derived Articles—Peptide Mapping  1055
1055 ).
).
Solution A:
Acetonitrile, water, and Sulfate buffer (100:700:200). Filter, and degas.
Solution B:
Acetonitrile, water, and Sulfate buffer (400:400:200). Filter, and degas.
Mobile phase:
See Table 3.
Table 3
| Time (min) |
Solution A (%) |
Solution B (%) |
|---|---|---|
| 0 | 95 | 5 |
| 3 | 95 | 5 |
| 30 | 41 | 59 |
| 35 | 20 | 80 |
| 40 | 95 | 5 |
| 50 | 95 | 5 |
Chromatographic system
Mode:
LC
Detector:
UV 214 nm
Column:
4.6-mm × 10-cm; 5-µm packing L1
Column temperature:
40
Flow rate:
1 mL/min
Injection volume:
50–100 µL
System suitability
Sample:
Standard solution
Suitability requirements
Chromatogram comparability:
In the chromatogram obtained from the Standard solution, identify the peaks due to digest fragments I, II, III, and IV. The chromatogram of the Standard solution corresponds to that of the typical chromatogram provided with the appropriate USP Insulin Reference Standard.
Tailing factor:
NMT 1.5
Resolution:
There should be complete separation of the peaks due to fragments II and III. The resolution is defined in the applicable insulin monograph.
Analysis
Samples:
Standard solution and Sample solution
Condition the Chromatographic system by running at initial conditions, t = 0 min, for at least 15 min. Carry out a blank gradient program before injecting the digests. Separately inject equal volumes of the Standard solution and the Sample solution, and record the responses of each peak.
Acceptance criteria:
The chromatographic profile of the Sample solution corresponds to that of the Standard solution.
LIMIT OF HIGH MOLECULAR WEIGHT PROTEINS
• Procedure
Solution A:
1 mg/mL of l-arginine in water
Mobile phase:
Solution A, acetonitrile, and glacial acetic acid (65:20:15). Filter, and degas.
Resolution solution:
Store a suitable amount of insulin drug substance at room temperature for a sufficient period of time (5–10 days, or as needed) to obtain insulin with more than 0.4% high molecular weight proteins. Prepare a 4-mg/mL solution in 0.01 N hydrochloric acid. Store the solution in a refrigerator, and use within 7 days. Alternatively, dissolve about 4 mg of USP High Molecular Weight Insulin Human RS in 1 mL of 0.01 N hydrochloric acid.
Sample solution:
In a small vial, prepare a 4-mg/mL solution of insulin in 0.01 N hydrochloric acid, and mix to dissolve. Store in a refrigerator, and use within 7 days.
Chromatographic system
Mode:
LC
Detector:
UV 276 nm
Column:
7.8-mm × 30-cm; 5- to 10-µm packing L20
Temperatures
Column:
Ambient
Autosampler:
It is advisable to use a refrigerated autosampler.
Flow rate:
0.5 mL/min
Injection volume:
100 µL
System suitability
Sample:
Resolution solution
Suitability requirements
Retention times:
Between 13 and 17 min for the polymeric insulin complexes, about 17.5 min for the covalent insulin dimer, and between 18 and 22 min for the insulin monomer, with salts eluting after the insulin monomer
Peak-to-valley ratio:
The ratio of the height of the covalent insulin dimer peak to the height of the valley between the covalent insulin dimer peak and the insulin monomer peak is NLT 2.0.
Analysis
Sample:
Sample solution
Disregard any peaks having retention times greater than that of the insulin monomer.
Calculate the percentage of high molecular weight proteins in the portion of Insulin taken:
Result = 100 × SrH/(SrH + rM)
| SrH | = | = sum of the responses of all peaks having retention times less than that of the insulin monomer |
| rM | = | = peak response of the insulin monomer |
Acceptance criteria:
As stated in the applicable insulin monograph
ADDITIONAL REQUIREMENTS
• USP Reference Standards  11
11
USP High Molecular Weight Insulin Human RS (alternative, optional)
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| General Chapter | Mary D. Crivellone, Ph.D.
Senior Reference Standards Scientist (301) 816-8156 |
(BIO12010) Monographs - Biologics and Biotechnology 1 |
| Reference Standards | RS Technical Services 1-301-816-8129 rstech@usp.org |
USP38–NF33 Page 195
Pharmacopeial Forum: Volume No. 39(3)