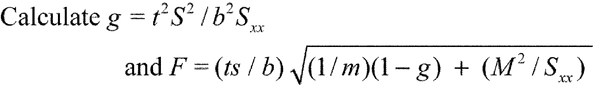Glucagon
(gloo' ka gon).
» Glucagon is a polypeptide hormone that has the property of increasing the concentration of glucose in the blood. It is obtained from porcine and bovine pancreas glands.
Packaging and storage—
Preserve in tight glass containers, under nitrogen, in a refrigerator.
Identification—
The retention time of the major peak in the chromatogram of the Test solution corresponds to that in the chromatogram of the Standard solution, as obtained in the test for Chromatographic purity.
Water, Method I  921
921 :
not more than 10.0%.
:
not more than 10.0%.
Residue on ignition  281
281 :
not more than 2.5%.
:
not more than 2.5%.
Chromatographic purity—
Phosphate–cysteine buffer solution—
Dissolve 18.9 g of monobasic sodium phosphate and 0.327 g of l-cysteine in 970 mL of water, adjust with phosphoric acid to a pH of 2.6, and dilute with water to 1000 mL.
Sample solvent—
Transfer 20.0 mL of acetonitrile to a 100-mL volumetric flask, and dilute with 0.01 N hydrochloric acid to volume.
Mobile phase—
Mix 270 mL of acetonitrile and 730 mL of Phosphate–cysteine buffer solution, and degas.
Standard solution—
Dissolve an accurately weighed quantity of USP Glucagon RS in Sample solvent to obtain a solution having a known concentration of about 0.5 mg per mL.
System suitability solution—
Dissolve an accurately weighed quantity of USP Glucagon RS in Sample solvent to obtain a solution containing about 0.5 mg per mL. Heat the solution at 75 for at least 3 hours to allow the formation of related substance GLU (24) Glucagon.
for at least 3 hours to allow the formation of related substance GLU (24) Glucagon.
Test solution—
Transfer about 12.5 mg of Glucagon, accurately weighed, to a 25-mL volumetric flask, and dissolve in Sample solvent to obtain a solution containing about 0.5 mg per mL.
Chromatographic system (see Chromatography  621
621 )—
The liquid chromatograph is equipped with a 214-nm detector and a 4.6-mm × 25-cm column that contains 5-µm packing L7. The column temperature is maintained at 35
)—
The liquid chromatograph is equipped with a 214-nm detector and a 4.6-mm × 25-cm column that contains 5-µm packing L7. The column temperature is maintained at 35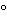 , and the flow rate is about 1.0 mL per minute. Chromatograph the System suitability solution, and record the peak responses as directed for Procedure: the resolution, R, between the main peak and the large peak with a relative retention time of 1.3 is not less than 4.0; and the tailing factor for the glucagon peak is not greater than 1.7.
, and the flow rate is about 1.0 mL per minute. Chromatograph the System suitability solution, and record the peak responses as directed for Procedure: the resolution, R, between the main peak and the large peak with a relative retention time of 1.3 is not less than 4.0; and the tailing factor for the glucagon peak is not greater than 1.7.
Procedure—
Separately inject equal volumes (about 50 µL) of the Standard solution and the Test solution into the chromatograph, record the chromatograms, and measure the peak responses. Calculate the percentage of each impurity in the portion of Glucagon taken by the formula:
100(ri / rs)
in which ri is the peak response for each individual impurity obtained from the Test solution; and rs is the sum of the responses of all the peaks: not more than 2.5% of any individual impurity is found; and not more than 10.0% of total impurities is found.
Nitrogen content, Method II  461
461 :
between 16.0% and 18.5%, calculated on the anhydrous basis.
:
between 16.0% and 18.5%, calculated on the anhydrous basis.
Zinc content  591
591 :
not more than 0.05%.
:
not more than 0.05%.
Assay—
[note—All buffers have a final pH of 7.4, unless otherwise indicated. The concentration range of the Standard preparations and the Assay preparations may be modified to fall within the linear range of the Assay. The calculations should be adjusted accordingly. Alternatively, full curve analysis using validated nonlinear statistical methods can be used, provided that similarity is demonstrated when comparing the responses of the Standard preparations and the Assay preparations.]
hepatocyte preparation—
Calcium-free perfusion buffer with dextrose—
Prepare a solution containing, in each L, 7.92 g of sodium chloride, 0.35 g of potassium chloride, 1.80 g of dextrose, 0.19 g of edetic acid, and 2.38 g of N-2-hydroxyethylpiperazine-N¢-2-ethanesulfonic acid. Oxygenate prior to circulation.
Collagenase buffer—
Prepare a solution containing, in each L, 3.62 g of sodium chloride, 23.83 g of N-2-hydroxyethylpiperazine-N¢-2-ethanesulfonic acid, 0.35 g of potassium chloride, 0.52 g of calcium chloride, and 1.8 g of dextrose. Adjust to a pH of 7.6, and oxygenate. Immediately before perfusion, dissolve a quantity of collagenase in this solution to obtain a concentration of 0.02% to 0.05%.
Wash buffer—
Prepare a solution containing, in each L, 7.92 g of sodium chloride, 0.35 g of potassium chloride, 0.19 g of edetic acid, 2.38 g of N-2-hydroxyethylpiperazine-N¢-2-ethanesulfonic acid, 0.22 g of calcium chloride, and 0.12 g of magnesium sulfate.
Incubation buffer—
Prepare a solution containing, in each L, 6.19 g of sodium chloride, 0.35 g of potassium chloride, 0.22 g of calcium chloride, 0.12 g of magnesium sulfate, 0.16 g of monobasic potassium phosphate, 11.915 g of N-2-hydroxyethylpiperazine-N¢-2-ethanesulfonic acid, and 1% bovine serum albumin (BSA). Adjust to a pH of 7.5.
Test animals—
Male Sprague-Dawley rats are maintained on a standard rat chow diet and freely given water. On the morning of the test, select a healthy rat weighing approximately 300 g, and administer 100 Units of Heparin Sodium subcutaneously.
Procedure—
[note—Conduct this procedure in the morning to ensure that the rat has optimal glycogen in its liver. ] Anesthetize the rat with an appropriate anesthetic. Open the abdominal cavity, and isolate the portal vein. Insert an angiocatheter connected to a perfusion pump, and tie into the portal vein at the general location of the lienal branch. Start the perfusion (25 mL per minute) in situ with Calcium-free perfusion buffer with dextrose, equilibrated with oxygen, at a temperature of 37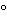 . As the liver enlarges, cut the inferior vena cava to allow pressure equilibrium. [note—About 300 mL of the perfusate is needed to clear the liver of red blood cells at a flow rate of 30 to 60 mL per minute. ] Then circulate Collagenase buffer at a flow rate of 30 to 60 mL per minute for about 10 minutes. The exact concentration of collagenase (within the range of 0.02% to 0.05%) is determined empirically for each lot of enzyme. The concentration of collagenase is that necessary to consistently cause a breakdown of the liver about 10 minutes after initial entry of the Collagenase buffer into the liver. When the liver significantly increases in size, changes color and consistency, and starts to leak perfusate out of the lobes, change the system to the oxygenated prewarmed Wash buffer. About 100 mL of Wash buffer is needed to wash the liver of collagenase at a flow rate of 25 mL per minute. Surgically remove the liver from the animal and place in a prewarmed tray containing oxygenated Wash buffer (37
. As the liver enlarges, cut the inferior vena cava to allow pressure equilibrium. [note—About 300 mL of the perfusate is needed to clear the liver of red blood cells at a flow rate of 30 to 60 mL per minute. ] Then circulate Collagenase buffer at a flow rate of 30 to 60 mL per minute for about 10 minutes. The exact concentration of collagenase (within the range of 0.02% to 0.05%) is determined empirically for each lot of enzyme. The concentration of collagenase is that necessary to consistently cause a breakdown of the liver about 10 minutes after initial entry of the Collagenase buffer into the liver. When the liver significantly increases in size, changes color and consistency, and starts to leak perfusate out of the lobes, change the system to the oxygenated prewarmed Wash buffer. About 100 mL of Wash buffer is needed to wash the liver of collagenase at a flow rate of 25 mL per minute. Surgically remove the liver from the animal and place in a prewarmed tray containing oxygenated Wash buffer (37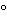 ). Gently comb the liver with a stainless steel, fine-toothed comb to free the hepatocytes. Wash the hepatocytes with Wash buffer, and filter through cheesecloth (or a 150-µm mesh polyethylene net) into a plastic beaker. Centrifuge the cell suspension for about 2 minutes at about 25 × g to form a loosely packed pellet. Discard the supernatant, and resuspend the pellet in Wash buffer. Repeat the washing procedure twice for a total of three washes. Resuspend the final pellet in 100 to 200 mL of Incubation buffer, depending on cell yield. [note—If the Assay procedure is interrupted, cool the cells by collecting the cells in a beaker placed in ice. The cells are washed with ice-cold Wash buffer, and stored on ice until ready for use. At that point the cells are pelleted once more, and resuspended in ice-cold Incubation buffer. ]
). Gently comb the liver with a stainless steel, fine-toothed comb to free the hepatocytes. Wash the hepatocytes with Wash buffer, and filter through cheesecloth (or a 150-µm mesh polyethylene net) into a plastic beaker. Centrifuge the cell suspension for about 2 minutes at about 25 × g to form a loosely packed pellet. Discard the supernatant, and resuspend the pellet in Wash buffer. Repeat the washing procedure twice for a total of three washes. Resuspend the final pellet in 100 to 200 mL of Incubation buffer, depending on cell yield. [note—If the Assay procedure is interrupted, cool the cells by collecting the cells in a beaker placed in ice. The cells are washed with ice-cold Wash buffer, and stored on ice until ready for use. At that point the cells are pelleted once more, and resuspended in ice-cold Incubation buffer. ]
Suitability—
The concentrations of cells may vary due to the collagenase activity and the viability of the hepatocytes. To check cell viability and to determine viable cell concentration, dilute duplicate 100-µL aliquots of cell suspension with 400 µL of Wash buffer and 500 µL of isotonic 0.4% trypan blue. The aliquots are counted in a hemocytometer. The cells are suspended in Incubation buffer to obtain a viable cell concentration of not less than 3 × 106 per mL. Count several distinct fields. [note—Viable cells are those cells that exclude the trypan blue. ]
negative control solution—
Prepare a solution containing 0.5% BSA in sterile water.
incubation flasks—
Use 25-mL conical flasks, the bottoms of which have been heated and pushed inward to form a conically raised center.
standard preparations—
In duplicate, dissolve a suitable quantity of USP Glucagon RS, accurately measured, in 0.01 N hydrochloric acid or other suitable diluent to obtain a solution containing 1.0 USP Glucagon Unit per mL. All dilutions thereafter are made using 0.5% BSA (w/v) in water. Accurately dilute measured volumes of each solution with Negative control solution to obtain five concentrations—200, 100, 50, 25, and 12.5 micro-Units per mL—of each solution (Standard preparations). Pipet 0.2 mL of each Standard preparation into separate Incubation flasks. Pipet 0.2 mL of Negative control solution into each of two flasks (Negative control solutions 1 and 2). Then add the hepatocytes into each of the 12 flasks.
assay preparations—
Using accurately weighed quantities of Glucagon, proceed as directed for Standard preparations.
d-glucose determination—
Standard stock solution—
Transfer 2.0 g of USP Dextrose RS, accurately weighed, to a 200-mL volumetric flask, and dissolve in and dilute with saturated benzoic acid solution to volume.
Standard solutions—
Transfer suitable quantities of Standard stock solution to three flasks, and dilute with saturated benzoic acid solution to obtain solutions having known concentrations of 0.5, 1.0, and 1.5 times the typical sample glucose concentration.
Potassium ferrocyanide solution—
Dissolve 1.25 g of trihydrate potassium ferrocyanide in 125 mL of Sterile Water for Injection.
System suitability—
Analyze the Potassium ferrocyanide solution, the Standard solutions, and five replicates of the middle Standard solution. Prepare a standard curve using the Standard solutions as directed for Procedure: the relative standard deviation of the standard curve is not more than 2.0%; the response of the Potassium ferrocyanide solution is not more than 30 mg per L; and the relative standard deviation is not more than 2.0% for the replicate analyses of the middle Standard solution.
procedure—
Dispense 5 mL of Hepatocyte preparation into the special incubation flasks in sequence from high glucagon concentration to low glucagon concentration, alternating the Standard preparations with the Assay preparations. The flasks are swirled in an orbiting water bath at 125 rpm at 30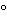 for approximately 30 to 60 minutes. [note—The exact incubation time must be determined to optimize the signal-to-noise ratio. ] Following incubation, place 0.5- to 1.0-mL aliquots, in duplicate, from each incubation flask into labeled tubes, and centrifuge at 12,500 × g. Determine the percentage of glucose concentration in each flask's supernatant.
for approximately 30 to 60 minutes. [note—The exact incubation time must be determined to optimize the signal-to-noise ratio. ] Following incubation, place 0.5- to 1.0-mL aliquots, in duplicate, from each incubation flask into labeled tubes, and centrifuge at 12,500 × g. Determine the percentage of glucose concentration in each flask's supernatant.
To conform to the linear range of the instrument being used, it may be necessary to adjust by dilution each of the preparations. Use a glucose analyzer that has demonstrated appropriate specificity, accuracy, precision, and linear response over the range of concentrations being determined. [note—A suitable analyzer may use an immobilized, oxidase-enzyme membrane or jacket-generating hydrogen peroxide, which is then detected at the electrode. ] Perform the glucose analysis in the following sequence: Negative control solution 1, Standard preparations, Assay preparations, and Negative control solution 2. Determine the percentage of glucose against the Negative control solution for each preparation.
calculations—
Linearity test—
Use an analysis of variance (ANOVA) with one sample assayed against a standard, and using two replicates each, construct a table (see Table 1). Compare the value of the ratio MSNL/MSRES1 to a critical value obtained from a table for an F distribution with m –2 and 3m –3 degrees of freedom, where m is the number of dose levels for each preparation. If the ratio MSNL/MSRES1 does not indicate the presence of significant nonlinearity (ratio value is lower than the critical value), then proceed to the test for parallelism. If the ratio exceeds the critical value (significance level of 0.025), the nonlinearity is statistically significant and the test is repeated, discarding the results from either the highest or lowest dose of both the Standard preparations and the Assay preparations (four dose levels). If the ratio MSNL/MSRES1 does not indicate the presence of significant nonlinearity, then proceed to the test for parallelism.
Parallelism test—
Compare the ratio MSNP/MSRES2 to a critical value obtained from an F distribution having 1 and 4m –5 degrees of freedom. If the ratio MSNP/MSRES2 does not indicate the presence of significant nonparallelism, then the assay is considered valid. Use the appropriate dose levels for the estimation of the relative potency.
Relative potency—
Calculate the relative potency, R, of the Assay preparations as compared with the Standard preparations as follows.
(1) Xj is defined as the log10 of the jth dose of the Standard preparations or the Assay preparations. The glucagon dose varies from 12.5 to 200 × 10–6 USP Glucagon Units per mL. For ease in the subsequent calculations, these doses are respectively represented by 1 through 5, as shown in the table below.
Xj is defined as the log10 of the jth dose of the Standard preparations or the Assay preparations. The glucagon dose varies from 12.5 to 200 × 10–6 USP Glucagon Units per mL. For ease in the subsequent calculations, these doses are respectively represented by 1 through 5, as shown in the table below.
| j
Dose |
1 12.5 |
2 25 |
3 50 |
4 100 |
5 200 |
|---|---|---|---|---|---|
| Xj | 1.10 | 1.40 | 1.70 | 2.00 | 2.30 |
(2) To differentiate between the Standard preparations and the Assay preparations in the calculations, the subscript i will be used, with i = 1 to designate the Standard preparations and i = 2 to designate the Assay preparations. Yijk will denote the glucose concentration associated with the kth replicate of the jth dose of the ith preparation. For example, Y1jk is the glucose concentration associated with the kth replicate of the jth dose of the appropriate Standard preparation; Y11k is the glucose concentration associated with the kth replicate of dose 1 of the Standard preparation; and Y21k is the glucose concentration associated with the kth replicate of dose 1 of the Assay preparation. Dose 1 represents a glucose dose of 12.5 × 10
To differentiate between the Standard preparations and the Assay preparations in the calculations, the subscript i will be used, with i = 1 to designate the Standard preparations and i = 2 to designate the Assay preparations. Yijk will denote the glucose concentration associated with the kth replicate of the jth dose of the ith preparation. For example, Y1jk is the glucose concentration associated with the kth replicate of the jth dose of the appropriate Standard preparation; Y11k is the glucose concentration associated with the kth replicate of dose 1 of the Standard preparation; and Y21k is the glucose concentration associated with the kth replicate of dose 1 of the Assay preparation. Dose 1 represents a glucose dose of 12.5 × 10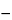 6 USP Glucagon Units per mL. Finally, Y132 represents the glucose concentration associated with the 2nd replicate of dose 3 for the Standard preparation.
6 USP Glucagon Units per mL. Finally, Y132 represents the glucose concentration associated with the 2nd replicate of dose 3 for the Standard preparation.
(3) YS and Yt denote the average glucose concentrations for the Standard preparations and the Assay preparations, respectively.
YS and Yt denote the average glucose concentrations for the Standard preparations and the Assay preparations, respectively.
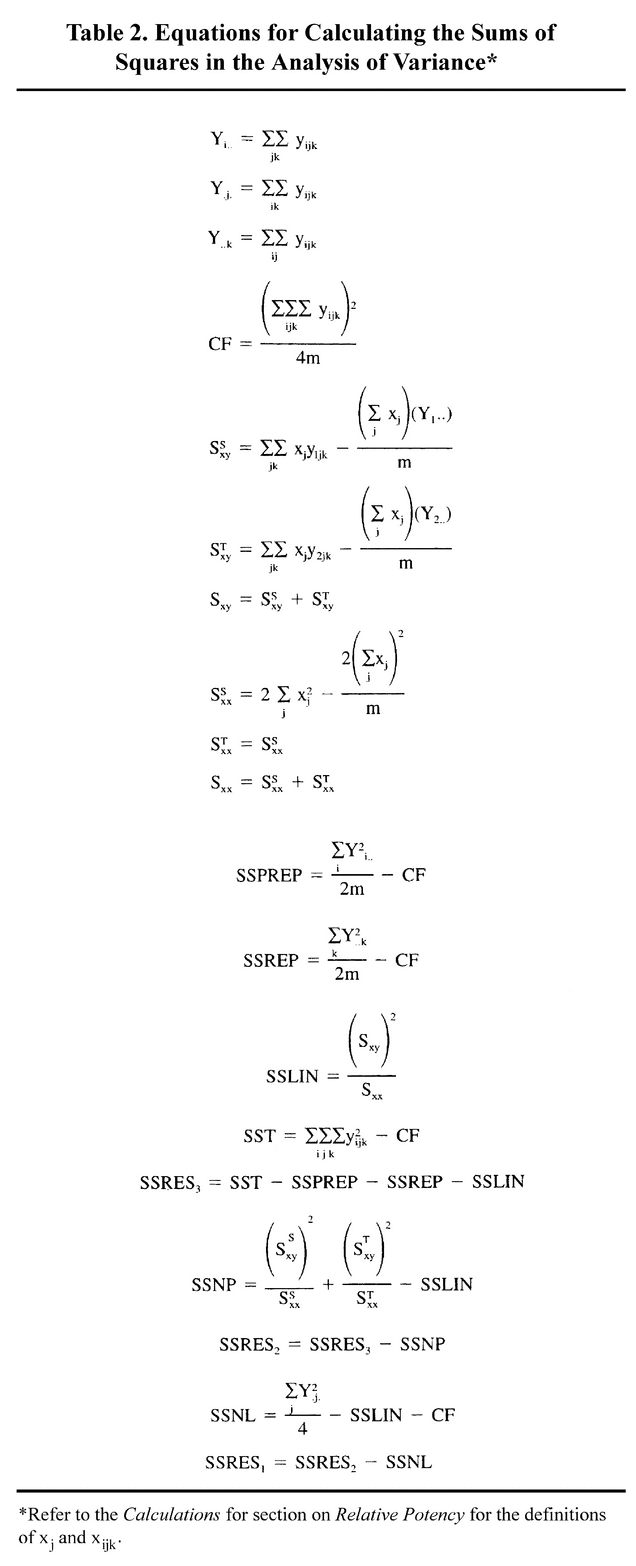
(4) Calculate the least-squares slope estimate, b, for a linear regression relating the Yijk's to the Xj's as follows: b = Sxy /Sxx , with Sxy and Sxx calculated using the equations in Table 2.
Calculate the least-squares slope estimate, b, for a linear regression relating the Yijk's to the Xj's as follows: b = Sxy /Sxx , with Sxy and Sxx calculated using the equations in Table 2.
(5) The log potency, M, is calculated using M =
The log potency, M, is calculated using M = 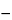 1[(YS
1[(YS 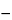 Y t )/b].
Y t )/b].
(6) R = antilog (M).
R = antilog (M).
(7) Calculate the confidence limits (upper and lower) for the relative potency, R, using the value s2 = MSRES3 (see Table 1 and Table 2) as follows. Obtain t from a table for a t distribution having 4m
Calculate the confidence limits (upper and lower) for the relative potency, R, using the value s2 = MSRES3 (see Table 1 and Table 2) as follows. Obtain t from a table for a t distribution having 4m 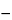 4 degrees of freedom. For the 95% limits, the t values can be obtained from Table 9 under Design and Analysis of Biological Assays
4 degrees of freedom. For the 95% limits, the t values can be obtained from Table 9 under Design and Analysis of Biological Assays  111
111 .
.
[note—For confidence limits having other probability levels (i.e., 100(1 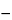 a) %), the right tail t critical value having a/2 area to its right is used. ]
a) %), the right tail t critical value having a/2 area to its right is used. ]
and calculate
ML = ( M 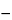 F)/(1
F)/(1 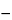 g)
g)
and
MU = ( M + F)/(1 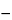 g)
g)
where M is the log potency and ML and MU are the log potency lower and upper confidence limits. The lower and upper confidence limits for the relative potency, R, are given by
RL = antilog ( ML)
RU = antilog ( MU)
It meets the requirements if the potency is between 0.8 and 1.25 USP Glucagon Units per mg, and the confidence interval width at P = 0.95 does not exceed 45% of the computed potency. Repeat the assay if the confidence interval width exceeds 45% of the computed potency.
Table 1. ANOVA for the Rat Hepatocyte Assay for Glucagon
ANOVA for the Rat Hepatocyte Assay for Glucagon
| Source | Degrees of Freedom | SS (Sum of Squares) | MS (Mean Square) |
|---|---|---|---|
| Preparations | 1 | SSPREP | MSPREP |
| Replicates | 1 | SSREP | MSREP |
| Linear Slope | 1 | SSLIN | MSLIN |
| Residual3 | 4m –4 | SSRES3 | MSRES3 |
| Nonparallelism | 1 | SSNP | MSNP |
| Residual2 | 4m –5 | SSRES2 | MSRES2 |
| Nonlinearity | m –2 | SSNL | MSNL |
| Residual1 | 3m –3 | SSRES1 | MSRES1 |
| TOTAL | 4m –1 | SST |
[notes—This analysis pertains to one sample assayed against a standard, using two replicates each. ]
The number of dose levels for each preparation is denoted by m.
Table 2 gives the equations for calculating the SS terms.
In each row of the ANOVA table, the MS is obtained by dividing the SS term by the degrees of freedom.
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| Monograph | Thomas A. Sigambris, M.S.
Scientific Liaison 1-301-998-6789 |
(BIO12010) Monographs - Biologics and Biotechnology 1 |
| Reference Standards | RS Technical Services 1-301-816-8129 rstech@usp.org |
USP35–NF30 Page 3342
Pharmacopeial Forum: Volume No. 37(4)