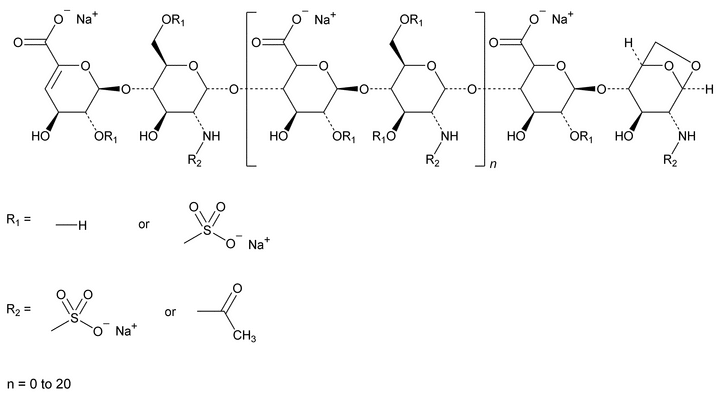
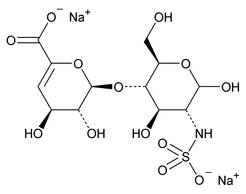
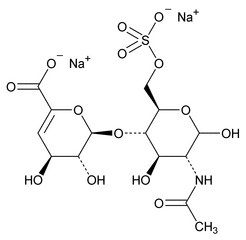
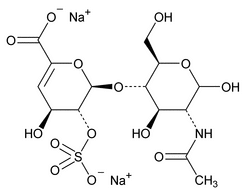
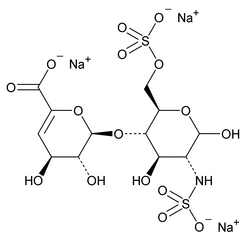
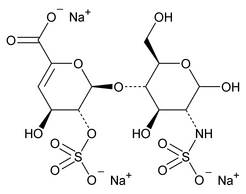
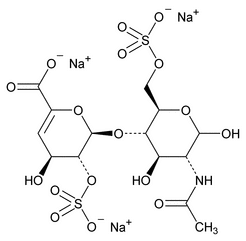
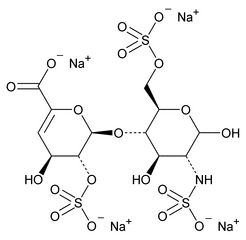
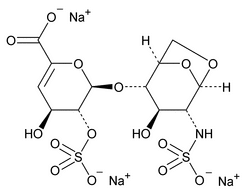
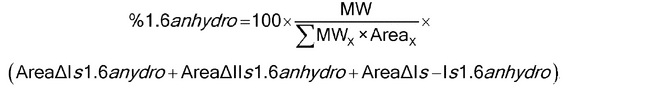The following procedure is used to determine the levels of 1, 6-anhydro forms in enoxaparin sodium. [Note—The test for the 1,6-anhydro derivative is conducted only where specified in the individual monograph. ]
INTRODUCTION
The disaccharides specified in this general chapter are listed by name and structure in Appendix 1; the oligosaccharides are listed in Appendix 2.
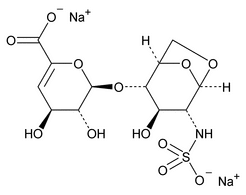
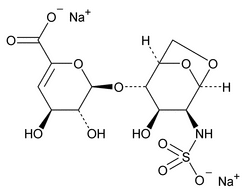
Depolymerization of heparin into enoxaparin sodium produces a partial but characteristic conversion of glucosamines at the reducing termini of oligosaccharide chains with terminal glucosamine 6-O sulfate, yielding 1,6-anhydro derivatives (see Figure 1).
Figure 1. Structure of enoxaparin sodium containing a 1,6-anhydro derivative on the reducing end of the chain.
The percentage of oligosaccharide chains that are cyclized in a 1,6-anhydro ring is a characteristic of enoxaparin sodium.
DEPOLYMERIZATION OF ENOXAPARIN SODIUM BY HEPARINASES AND RESULTING OLIGOSACCHARIDES
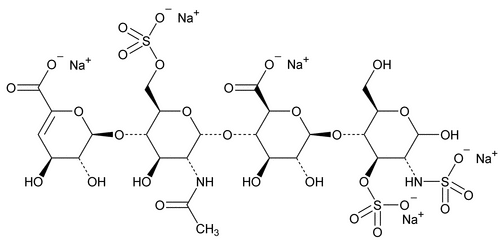
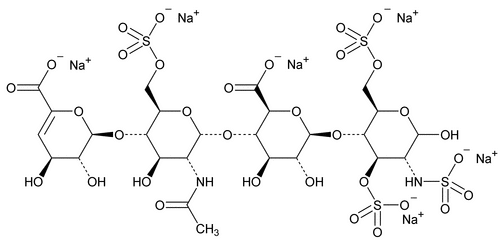
The assay involves HPLC analysis of a depolymerized enoxaparin sodium solution by a mixture of heparinases. After enzymatic depolymerization, the main 1,6-anhydro residues of enoxaparin sodium observed are 1,6-anhydro DIIS and 1,6-anhydro DIISepi, and 1,6-anhydro DIS and 1,6-anhydro DIS–ISepi (see Appendix 2.).
The 1,6-anhydro DIS–ISepi tetrasaccharide (2-O-sulfated mannosamine form) is not completely cleaved by the heparinases. The two disaccharides (1,6-anhydro DIIS and 1,6-anhydro DIISepi), which generally co-elute, are poorly resolved with respect to DIIA (see Appendix 1), especially because the latter occurs as two anomers: 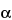 and
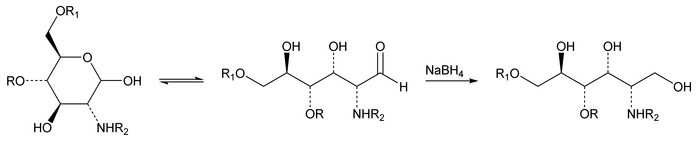
and 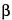 . To allow quantitation of 1,6-anhydro DIIS and 1,6-anhydro DIISepi, the enoxaparin sodium sample already depolymerized by heparinases is then reduced by sodium borohydride (see Figure 2).
. To allow quantitation of 1,6-anhydro DIIS and 1,6-anhydro DIISepi, the enoxaparin sodium sample already depolymerized by heparinases is then reduced by sodium borohydride (see Figure 2).
The sodium borohydride reduction eliminates the  anomeric effect by opening the terminal oligosaccharide ring. The four 1,6-anhydro derivatives (see Appendix 2.) are not reduced by sodium borohydride because the ring opening is blocked by the 1,6-anhydro bridge. The reduction of the oligosaccharides decreases their retention time, whereas the retention time of the 1,6-anhydro derivatives remains unchanged. Thus, it is possible to separate the two compounds—1,6-anhydro DIIS and 1,6-anhydro DIISepi—from the reduced DIIA disaccharide peak. [Note—1,6-Anhydro DIIS and 1,6-anhydro DIISepi are eluted as two nonresolved peaks and are quantitated together as a single compound, 1,6-anhydro DIIS. Therefore, for the purpose of simplification, the epimeric form is not referred to in the remaining text. ]
anomeric effect by opening the terminal oligosaccharide ring. The four 1,6-anhydro derivatives (see Appendix 2.) are not reduced by sodium borohydride because the ring opening is blocked by the 1,6-anhydro bridge. The reduction of the oligosaccharides decreases their retention time, whereas the retention time of the 1,6-anhydro derivatives remains unchanged. Thus, it is possible to separate the two compounds—1,6-anhydro DIIS and 1,6-anhydro DIISepi—from the reduced DIIA disaccharide peak. [Note—1,6-Anhydro DIIS and 1,6-anhydro DIISepi are eluted as two nonresolved peaks and are quantitated together as a single compound, 1,6-anhydro DIIS. Therefore, for the purpose of simplification, the epimeric form is not referred to in the remaining text. ]
 anomeric effect by opening the terminal oligosaccharide ring. The four 1,6-anhydro derivatives (see Appendix 2.) are not reduced by sodium borohydride because the ring opening is blocked by the 1,6-anhydro bridge. The reduction of the oligosaccharides decreases their retention time, whereas the retention time of the 1,6-anhydro derivatives remains unchanged. Thus, it is possible to separate the two compounds—1,6-anhydro DIIS and 1,6-anhydro DIISepi—from the reduced DIIA disaccharide peak. [Note—1,6-Anhydro DIIS and 1,6-anhydro DIISepi are eluted as two nonresolved peaks and are quantitated together as a single compound, 1,6-anhydro DIIS. Therefore, for the purpose of simplification, the epimeric form is not referred to in the remaining text. ]
anomeric effect by opening the terminal oligosaccharide ring. The four 1,6-anhydro derivatives (see Appendix 2.) are not reduced by sodium borohydride because the ring opening is blocked by the 1,6-anhydro bridge. The reduction of the oligosaccharides decreases their retention time, whereas the retention time of the 1,6-anhydro derivatives remains unchanged. Thus, it is possible to separate the two compounds—1,6-anhydro DIIS and 1,6-anhydro DIISepi—from the reduced DIIA disaccharide peak. [Note—1,6-Anhydro DIIS and 1,6-anhydro DIISepi are eluted as two nonresolved peaks and are quantitated together as a single compound, 1,6-anhydro DIIS. Therefore, for the purpose of simplification, the epimeric form is not referred to in the remaining text. ]
PROCEDURE
USP Reference Standards
 11
11 —USP Enoxaparin Sodium RS.
—USP Enoxaparin Sodium RS.
Solutions—
Solution A—
Dissolve 0.280 g of monobasic sodium phosphate in 950 mL of water, adjust with phosphoric acid to a pH of 3.0, and dilute with water to 1000 mL.
Solution B—
Dissolve 140 g of sodium perchlorate in 950 mL of Solution A, adjust with phosphoric acid to a pH of 3.0, and dilute with Solution A to 1000 mL.
Mobile Phase—
Use variable mixtures of filtered and degassed Solution A and Solution B as directed in Chromatographic System.
Sodium/Calcium Acetate pH 7.0 Solution—
Dissolve 10 mg of bovine serum albumin and 32 mg of calcium acetate in 60 mL of water. Add 580 µL of glacial acetic acid, and adjust with 2 M sodium hydroxide to a pH of 7.0. Transfer to a 100-mL volumetric flask, and dilute with water to volume. Pass the solution through a filter having a porosity of 0.45 or 0.22 µm.
Potassium Phosphate pH 7.0 Buffer—
Dissolve 68 mg of monobasic potassium phosphate and 10 mg of bovine serum albumin in 30 mL of water in a 50-mL volumetric flask. Adjust with potassium hydroxide, if necessary, to a pH of 7.0, and dilute with water to volume. Pass the solution through a filter having a porosity of 0.45 or 0.22 µm.
Sodium Borohydride Solution—
Dissolve 12 mg of sodium borohydride in 400 µL of water, and mix on a vortex mixer. [Note—Prepare fresh immediately before use. ]
Heparinase 1 Solution—
Dissolve heparinase 1 (see Reagent Specifications under Reagents, Indicators, and Solutions) [reference: heparin lyase I, EC 4.2.2.7] in Potassium Phosphate pH 7.0 Buffer to obtain a solution having an activity of 0.4 IU per mL. Store the solution at –20 until ready to use. [Note—Heparinase solutions can be stored for 3 months at
until ready to use. [Note—Heparinase solutions can be stored for 3 months at  20
20 . ]
. ]
Heparinase 2 Solution—
Dissolve heparinase 2 (see Reagent Specifications under Reagents, Indicators, and Solutions [no EC number] in Potassium Phosphate pH 7.0 Buffer to obtain a solution having an activity of 0.4 IU per mL. Store the solution at –20 until ready to use.
until ready to use.
Heparinase 3 Solution—
Dissolve heparinase 3 (see Reagent Specifications under Reagents, Indicators, and Solutions [reference: heparitinase I, EC 4.2.2.8] in Potassium Phosphate pH 7.0 Buffer to obtain a solution having an activity of 0.4 IU per mL. Store the solution at –20 until ready to use.
until ready to use.
Heparinases 1, 2, 3, Solution—
Prepare a 1:1:1 (v:v:v) mixture of Heparinase 1 Solution, Heparinase 2 Solution, and Heparinase 3 Solution.
Peak Identification Solutions—
[Note—The depolymerized test solutions and Standard solutions must be prepared at the same time. Depolymerized test solutions are stable for 1 month at  20
20 . Also, the reduced test solutions and Standard solutions must be prepared at the same time. Reduced solutions are also stable for 1 month at
. Also, the reduced test solutions and Standard solutions must be prepared at the same time. Reduced solutions are also stable for 1 month at  20
20 . ]
. ]
Disaccharide Solutions—
Separately prepare a 0.25 mg per mL solution of each disaccharide1 DIA, DIIA, DIIIA, DIVA, DIS, DIIS, DIIIS, DIVS (see Appendix 1). Chromatograph each disaccharide solution, and record the peak responses.
Reduced Disaccharide Solutions—
To 60 µL of each Disaccharide Solution, add 10 µL of freshly prepared Sodium Borohydride Solution. Mix on a vortex mixer, and allow to stand at room temperature for at least 4 hours. Chromatograph each solution, and record the peak response.
Blank Solution—
Prepare a mixture of 20 µL of water, 70 µL of Sodium/Calcium Acetate pH 7.0 Solution, and 100 µL of the Heparinases 1, 2, 3 Solution. Mix gently by inversion, and allow to stand for at least 48 hours in a 25 water bath. Prepare a mixture of 60 µL of this depolymerized solution with 10 µL of freshly prepared Sodium Borohydride Solution. Homogenize, and allow to stand at room temperature for at least 4 hours. Chromatograph the resulting solution, and record the peak responses.
water bath. Prepare a mixture of 60 µL of this depolymerized solution with 10 µL of freshly prepared Sodium Borohydride Solution. Homogenize, and allow to stand at room temperature for at least 4 hours. Chromatograph the resulting solution, and record the peak responses.
Test Solution 1—
Prepare two solutions, each containing 20 mg of enoxaparin sodium in 1 mL of water.
Standard Solution 1—
Prepare one solution containing 20 mg of USP Enoxaparin Sodium RS in 1 mL of water.
Test Solution 2—
For each solution, prepare a mixture of 20 µL of Test Solution 1, 70 µL of Sodium/Calcium Acetate pH 7 Solution, and 100 µL of Heparinases 1, 2, 3 Solution. Mix gently by inversion, and allow to stand for at least 48 hours in a 25 water bath. After 48 hours of depolymerization, chromatograph the solution, and record the peak responses.
water bath. After 48 hours of depolymerization, chromatograph the solution, and record the peak responses.
Standard Solution 2—
Prepare a mixture of 20 µL of Standard Solution 1, 70 µL of Sodium/Calcium Acetate pH 7 Solution, and 100 µL of Heparinases 1, 2, 3 Solution. Mix gently, and allow to stand for at least 48 hours in a 25 water bath. After 48 hours of depolymerization, chromatograph the solution, and record the peak responses.
water bath. After 48 hours of depolymerization, chromatograph the solution, and record the peak responses.
Test Solution 3—
For each depolymerized test solution, prepare a mixture of 60 µL of Test Solution 2 and 10 µL of freshly prepared Sodium Borohydride Solution. Homogenize, and allow to stand loosely capped at room temperature for at least 4 hours before injecting into the chromatograph. Test Solution 3 is stable for 48 hours at room temperature.
Standard Solution 3—
Prepare a mixture of 60 µL of Standard Solution 2 and 10 µL of freshly prepared Sodium Borohydride Solution. Homogenize and mix on a vortex mixer, and allow to stand loosely capped at room temperature for at least 4 hours before injecting into the chromatograph. Standard Solution 3 is stable for 48 hours at room temperature.
Chromatographic System (see Chromatography  621
621 )—
The liquid chromatograph is equipped with a 234-nm detector and a 3-mm × 25-cm column that contains 5-µm packing L14. A guard column packed with the same material should also be used. The flow rate is 0.45 mL per minute, the column temperature is maintained at 50
)—
The liquid chromatograph is equipped with a 234-nm detector and a 3-mm × 25-cm column that contains 5-µm packing L14. A guard column packed with the same material should also be used. The flow rate is 0.45 mL per minute, the column temperature is maintained at 50 , and the injection volume is 10 µL. The chromatograph is programmed as follows.
, and the injection volume is 10 µL. The chromatograph is programmed as follows.
Chromatograph the reduced Test Solution 3 and the reduced Standard Solution 3, and record the peak responses as directed for Procedure.
| Time (minutes) |
Solution A (%) | Solution B (%) | Elution |
|---|---|---|---|
| 0–20 | 97®65 | 3®35 | Linear gradient |
| 20–50 | 65®0 | 35®100 | Linear gradient |
| 50–60 | 0 | 100 | Isocratic |
| 60–61 | 0®97 | 100®3 | Linear gradient for re-equilibration |
| 61–79 | 97 | 3 | Isocratic for re-equilibration |
Depolymerization Suitability Test—
The ratio of the peak area of 1,6-anhydro-DIS-IS to that of 1,6-anhydro DIS is not more than 1.15 for the depolymerized Standard Solution 2.
Column Performance Suitability Test—
Identify the peaks corresponding to reduced DIA and 1,6-anhydro-DIS for the Standard Solution 3: the retention time of reduced DIS is between 27 and 33 minutes for the depolymerized and reduced Standard Solution 3; and the resolution, R, between reduced DIA and 1,6-anhydro-DIS is not less than 1.5.
Reduction Suitability Test—
The ratio of the peak area of DIS disaccharide to that of reduced DIS in the depolymerized and reduced Standard Solution 3 and Test Solution 3 is not more than 0.02%.
Procedure/Calculation—
Separately inject equal volumes of the reduced Test Solutions 3 and the reduced Standard Solution 3 into the chromatograph. Use the normalized area percentage method for calculation. Each peak is integrated from the dwell volume peak to the last detected peak. Measure the area of each analyte peak after excluding solvent peaks at the beginning of the chromatogram and in the Blank Solution. Using the previously obtained chromatograms of the Reduced Disaccharide Solutions, identify peaks belonging to the eight reduced disaccharides in the chromatograms for Test Solution 3 and Standard Solution 3. The peaks belonging to 1,6-Anhydro DIS, 1,6-Anhydro DIIS, and 1,6-Anhydro DIS-ISepi are identified from the relative retention times provided in Table 1 and the Reference chromatogram provided with USP Enoxaparin Sodium RS. Once the peaks have been identified, use the values in Table 1 to calculate the (w/w) percentage of the three main 1,6-anhydro derivatives obtained after depolymerization of enoxaparin sodium using the following formula:
% 1,6-anhydro i (w/w) = (100 × MWi × Ai)/ S(MWx × Ax)
in which MWi and Ai are the molecular weight and the area of the 1,6-anhydro peak i, respectively; and MWx and Ax are the molecular weight and the area, respectively, of either the peak X or the zone X specified by its retention time. [Note—Once the method is established, the peaks belonging to the different di- and tetrasaccharides can be easily identified using the USP Enoxaparin Sodium RS chromatogram. Thus, the use of the disaccharide Standards is only needed during the method-implementation stage. ]
Calculate the molar percentage of components containing a 1,6-anhydro structure at the reducing end of their chain in the enoxaparin sodium test sample according to the following formula:
in which MW is the mass-average molecular mass (see Identification test D under Enoxaparin Sodium); MWx and Areax are the molecular weight and the area, respectively, of either the peak X or the range X specified by its retention time. The molar percentage of components having a 1,6-anhydro structure at the reducing end of their chain is between 15% and 25%. Typical retention times and molecular masses attributed to different oligosaccharide structures are provided in Table 1.
Table 1. Typical Relative Retention Times (tRR) and Molecular Masses Attributed to Different Compounds*
| Compound | tRR | Molecular Mass (Daltons) |
|---|---|---|
| — | < 0.25 | 741 |
| Reduced DIVA | 0.25 | 401 |
| — | 0.25 <tRR < 0.51 | 741 |
| Reduced DIVS | 0.51 | 461 |
| — | 0.51 < tRR < 0.55 | 483 |
| Reduced DIIA | 0.55 | 503 |
| — | 0.55 < tRR < 0.59 | 503 |
| 1,6-Anhydro DIIS | 0.59 | 443 |
| — | 0.59 < tRR < 0.64 | 503 |
| Reduced DIIIA | 0.64 | 503 |
| — | 0.64 < tRR < 0.72 | 533 |
| Reduced DIIS | 0.72 | 563 |
| — | 0.72 < tRR < 0.80 | 563 |
| Reduced DIIIS | 0.80 | 563 |
| — | 0.80 < tRR < 0.88 | 583 |
| Reduced DIA | 0.88 | 605 |
| — | 0.88 < tRR < 0.90 | 635 |
| 1,6-Anhydro DIS | 0.90 | 545 |
| — | 0.90 < tRR <0.98 | 635 |
| Reduced DIIA-IVSglu | 0.98 | 1066 |
| — | 0.98< tRR <1.00 | 635 |
| Reduced DIS | 1.00 | 665 |
| — | 1.00< tRR <1.04 | 665 |
| DIS | 1.04 | 665 |
| — | 1.04< tRR <1.10 | 1228 |
| Reduced DIIA-IISglu | 1.10 | 1168 |
| — | 1.10< tRR <1.27 | 1228 |
| 1,6-Anhydro DIS-IS | 1.27 | 1210 |
| — | tRR >1.27 | 1228 |
|
*
Relative retention times were obtained with a depolymerized and reduced batch of enoxaparin sodium. They are expressed relative to the retention time of the main peak corresponding to reduced DIS. Note that according to the quality of the column, relative retention times can change slightly.
|
||
APPENDIX 1: STANDARD DISACCHARIDE STRUCTURES
| DIVA | DUA-(1®4) |
|
| DIVS | DUA-(1®4) |
|
| DIIA | DUA-(1®4) |
|
| DIIIA | DUA-2S-(1®4) |
|
| DIIS | DUA-(1®4) |
|
| DIIIS | DUA-2S-(1®4) |
|
| DIA | DUA-2S-(1®4) |
|
| DIS | UA-2S-(1®4) |
1
Suitable disaccharides are available from Grampian Enzymes (GE-H1001, GE-G1002, GE-H1003, GE-H1004, GE-H1005, GE-H1006, GE-H1007, GE-H1008), Nisthouse, Harray, Orkney, KW17 2LQ, United Kingdom, Tel: 01856 771771, Scottish Local Authority: Orkney Islands.
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| General Chapter | Anita Y. Szajek, Ph.D.
Principal Scientific Liaison 1-301-816-8325 |
(BIO12010) Monographs - Biologics and Biotechnology 1 |
USP35–NF30 Page 136
Pharmacopeial Forum: Volume No. 34(1) Page 143