SCOPE
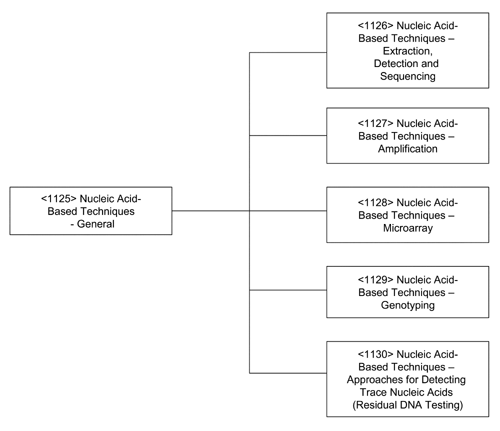
Nucleic acid-based assays are used in a variety of settings, the most common of which include the detection of infectious agents (viruses, bacteria, etc.), and cellular materials, as well as disease profiling. More recently such assays have also been used for forensic purposes and for the detection of trace contamination in biological materials. The latter include pharmaceutical development applications, such as viral clearance and adventitious agent testing in vaccine seed lots and tissue culture cell banks. This chapter introduces a series of general information chapters that provide techniques that support procedures for the detection and analysis of nucleic acids (see Figure 1). The assays using these techniques may be presented in a USP general chapter or in a private specification.
The major requirement for any nucleic acid analytical procedure is the availability of pure, intact nucleic acids for analysis. The information in Nucleic Acid-Based Techniques—Extraction, Detection, and Sequencing  1126
1126 discusses procedures available for nucleic acid extraction and handling. Hybridization is the core mechanism underlying many molecular biology techniques, and in addition to the detection of nucleic acids by absorbance and fluorescence measurements and size measurement by gel electrophoresis, this chapter also covers blotting and identification of nucleic acid species by hybridization assays using labeled probes. Hybridization probes are oligonucleotides that have a sequence that is complementary to the target of interest. Probes contain radioactive, fluorescent, biotin, digoxigenin, or other tags that, upon binding of the probe to the target, allow visualization and identification of the target. Probes are capable of detecting target sequences that are present in concentrations too low to be detected by absorbance measurements or gel electrophoresis.
discusses procedures available for nucleic acid extraction and handling. Hybridization is the core mechanism underlying many molecular biology techniques, and in addition to the detection of nucleic acids by absorbance and fluorescence measurements and size measurement by gel electrophoresis, this chapter also covers blotting and identification of nucleic acid species by hybridization assays using labeled probes. Hybridization probes are oligonucleotides that have a sequence that is complementary to the target of interest. Probes contain radioactive, fluorescent, biotin, digoxigenin, or other tags that, upon binding of the probe to the target, allow visualization and identification of the target. Probes are capable of detecting target sequences that are present in concentrations too low to be detected by absorbance measurements or gel electrophoresis.
These analytical procedures require a minimum quantity of nucleic acid, typically in the nanogram to microgram range. However, in the vast majority of cases, e.g., in the detection of viruses or rare cellular RNA species, the nucleic acid under assay is present in minute quantities (in the picogram to femtogram range), and an amplification step must be performed before the nucleic acid can be detected and identified. The amplification step may be directed either at the signal used for detection (signal amplification), such as the branched DNA assay (bDNA assay), or at the target as in nucleic acid amplification technologies (NAT).
In 1983 a revolutionary yet simple process termed polymerase chain reaction (PCR) was developed for amplifying the number of specific nucleic acid fragments present in a sample, and in just a few years after its discovery PCR became the most frequently used procedure for amplifying nucleic acids, especially DNA. Since the inception of PCR, the number of applications has expanded rapidly, and the technique, which now includes quantitative and multiplex assays, is currently used in almost every field of research and development in biology and medicine. Numerous variations of assay procedures have been developed for specific analytes. The general information chapter, Nucleic Acid-Based Techniques—Amplification  1127
1127 , describes amplification procedures used for DNA and RNA analysis as well as qualitative and quantitative NAT assays. Signal amplification procedures in which the signals, typically fluorescent signals, are used to detect the nucleic acid of interest, are not very common. The major signal amplification procedure, the branched DNA or bDNA assay, is used predominantly for viral nucleic acid detection.
, describes amplification procedures used for DNA and RNA analysis as well as qualitative and quantitative NAT assays. Signal amplification procedures in which the signals, typically fluorescent signals, are used to detect the nucleic acid of interest, are not very common. The major signal amplification procedure, the branched DNA or bDNA assay, is used predominantly for viral nucleic acid detection.
Quality assurance aspects of the methodology are also covered, together with a summary of current regulatory requirements for NAT assays. The need for globally comparable, accurate, and reliable results in the diagnostics field has driven the quest for, and development of, national and international standards within an increasingly sophisticated and metrologically sound, highly developed international regulatory environment devoted to the highest standards of regulatory science. Because NAT has become the most widely used of nucleic acid techniques, the majority of guidance documents and standards are related to NAT. The general information chapter, Nucleic Acid-Based Techniques—Microarrays  1128
1128 , addresses a still-emerging field that is of increasing relevance to molecular DNA analysis. Detailed treatment of various microarrays, including data analysis and validation, are excluded from
, addresses a still-emerging field that is of increasing relevance to molecular DNA analysis. Detailed treatment of various microarrays, including data analysis and validation, are excluded from  1128
1128 at this time. The general information chapter, Nucleic Acid-Based Techniques—Genotyping
at this time. The general information chapter, Nucleic Acid-Based Techniques—Genotyping  1129
1129 , focuses on the specific modifications of the techniques that are necessary to enable detection of single base differences and common genetic variations, e.g., single nucleotide polymorphisms (SNPs). The final general information chapter in the series, Nucleic Acid-Based Techniques—Approaches for Detecting Trace Nucleic Acids (Residual DNA Testing)
, focuses on the specific modifications of the techniques that are necessary to enable detection of single base differences and common genetic variations, e.g., single nucleotide polymorphisms (SNPs). The final general information chapter in the series, Nucleic Acid-Based Techniques—Approaches for Detecting Trace Nucleic Acids (Residual DNA Testing)  1130
1130 , describes residual DNA testing in the context of pharmaceutical manufacturing. Applications relevant to viral adventitious agents, however, are discussed in the general information chapter Virology Test Methods
, describes residual DNA testing in the context of pharmaceutical manufacturing. Applications relevant to viral adventitious agents, however, are discussed in the general information chapter Virology Test Methods  1237
1237 .
.
Two major uses of nucleic acid testing are excluded from this family of NAT chapters: viral testing for blood and blood product safety and genetic testing. The traditional perspective of USP is to develop public standards that can be applied to a particular final product without expressively defining a product and/or its production details. This chapter aims to specify when traditional methodologies or existing standards can be adapted. Novel methodologies for amplification and detection by NAT are also highlighted. As these new methodologies become mature and properly validated, they will be included in subsequent revisions.
Due to rapid development in the field, compendial and regulatory affairs scientists are advised to consult the current edition of USP and its Supplements regularly.
APPENDIX: REGULATIONS AND STANDARDS
Nucleic acid-based techniques have rapidly transformed almost every field of research, pharmaceutical development, and diagnostics. The need for globally comparable, accurate, and reliable results in the diagnostic field has driven the development of national and international standards as well as fostered a highly developed regulatory environment. Because NAT has become the most widely used of nucleic acid techniques, the majority of guidance documents and standards are related to NAT.1 Virus-specific regulations and reference standards will be addressed in the Appendix to General Information chapter Virology Test Methods  1237
1237 . The following is a selective list of national guidance documents. For application-specific guidance the compendial user is referred back to the relevant regulatory agency for the most current guidance.
. The following is a selective list of national guidance documents. For application-specific guidance the compendial user is referred back to the relevant regulatory agency for the most current guidance.
- FDA Center for Biologics Evaluation (CBER) “Review Criteria for Nucleic Acid Amplification-Based In Vitro Diagnostic Devices for Direct Detection of Infectious Microorganisms” (1993)
- FDA Center for Biologics Evaluation (CBER) “Guidance for Industry: Content and Format of Chemistry, Manufacturing and Controls Information and Establishment Description Information for a Biological In Vitro Diagnostic Product” (1999)
- FDA Center for Biologics Evaluation (CBER) “Guidance for Industry: In the Manufacture and Clinical Evaluation of In Vitro Tests to Detect Nucleic Acid Sequences of Human Immunodeficiency Viruses Types 1 and 2” (1999)
- FDA Center for Biologics Evaluation (CBER) “Guidance for Industry: Use of Nucleic Acid Tests on Pooled and Individual Samples from Donations of Whole Blood and Blood Components (including Source Plasma and Source Leukocytes) to Adequately and Appropriately Reduce the Risk of Transmission of HIV-1 and HCV” (2004)
GLOSSARY
3¢–5¢ exonuclease activity—
Enzymatic activity to remove a mispaired nucleotide from the 3¢ end of the growing strand. The reaction is a hydrolysis of a phosphoester bond. The presence of a 3¢–5¢ exonuclease, or proofreading, activity improves the fidelity of the polymerization.
5¢–3¢ exonuclease activity—
Enzymatic activity to remove a mispaired nucleotide from the 5¢ end of a polynucleotide strand. This activity is actually that of a single-strand-dependent endonuclease and is needed to remove RNA primers of Okazaki fragments, the RNA strand in the intermediate DNA–RNA heteroduplex during reverse transcription, and during DNA repair.
absorbance [Symbol: A]
—The logarithm, to the base 10, of the reciprocal of the transmittance (T). [note—Descriptive terms used formerly include optical density, absorbancy, and extinction.]
accuracy—
The accuracy of an analytical procedure is the closeness of test results obtained by that procedure to the true value.
allele—
One of two or more alternative forms of a gene at a given position (locus) on a chromosome, caused by a difference in the sequence of DNA.
amplicon—
A short segment of DNA generated by the PCR process whose sequence is defined by forward and reverse primers. Sometimes referred to as an amplimer.
amplification—
The enzymatic in vitro replication of a target nucleic acid.
annealing—
Hybridizing or binding of complementary nucleic acids, usually at an optimal temperature.
concatenation—
The process in which a DNA segment composed of repeated sequences is linked end-to-end.
complementary dna (cdna)—
DNA synthesized from an RNA template in an enzymatic reaction catalyzed by the enzyme reverse transcriptase.
denaturation—
The process of separating double-stranded DNA into single strands by breaking the hydrogen bonds. This is typically accomplished by heating the DNA solution to temperatures greater than 90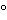 or by treating it with strong alkali.
or by treating it with strong alkali.
deoxyribonucleic acid (dna)—
The genetic material that is passed from parent to daughter cells and propagates the characteristics of the species in the form of genes it contains and the proteins for which it codes. DNA contains the following four deoxyribonucleosides: dA, dC, dT, and dG.
deoxyribonucleotide triphosphate (dntp)—
A base that is added to a primer during the PCR that comprises the newly synthesized strand. Examples of dNTPs are dATP, dUTP, dCTP, dGTP, and dTTP.
detection limit—
It is the lowest amount of analyte in a sample that can be detected, but not necessarily quantitated, under the stated experimental conditions.
dna polymerase—
An enzyme that can synthesize new complementary DNA strands using a DNA template and primer. Several of these enzymes are commercially available, including Taq DNA polymerase and rTth DNA polymerase.
endonuclease—
An enzyme that cleaves phosphodiester bonds in a polynucleotide chain.
energy transfer—
This describes the process in which an excited state of one molecular entity (the donor) is deactivated to a lower-lying state by transferring energy to a second molecular entity (the acceptor), which is thereby raised to a higher energy state.
extension—
Refers to the elongation of the DNA chain that is being synthesized using the parent DNA strand as the template for synthesis of that daughter strand. This is a natural process that occurs during DNA replication. Extension occurs during the PCR process with DNA polymerases.
extinction coefficient [Symbol: 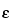 ]
—The quotient of the absorbance (A) divided by the product of the concentration, expressed in moles/L, of the substance and the absorption path length, in cm. [note—Terms formerly used include molar absorbancy index; molar absorptivity; and molar absorption coefficient.]
]
—The quotient of the absorbance (A) divided by the product of the concentration, expressed in moles/L, of the substance and the absorption path length, in cm. [note—Terms formerly used include molar absorbancy index; molar absorptivity; and molar absorption coefficient.]
fidelity—
Fidelity is a measure of the accuracy of nucleic acid replication. The polymerase enzyme used is only one of the elements that influences fidelity. Other elements include buffer conditions, thermal cycling parameters, number of cycles, efficiency of amplification, and the sequence of the DNA being copied.
fluorophore—
A functional group in a molecule that makes the molecule fluorescent by absorbing energy of a specific wavelength and re-emits the energy at another wavelength.
fluorescence—
The emission of one or more photons by a molecule or atom activated by the absorption of a quantum of electromagnetic radiation. X-rays, UV, visible light, and IR radiations may all stimulate fluorescence. For details on the spectroscopic measurement of fluorescence, see Spectrophotometry and Light-Scattering  851
851 .
.
genome—
The complete genetic complement or the complete set of instructions for reproducing an organism and carrying out its biological function in life. The DNA in our cells comprises our genome. When our cells divide, the complete genome in our cells is duplicated for transmission to each of the remaining daughter cells.
genotype—
The genetic constitution of an organism as revealed by genetic or molecular analysis, i.e., the complete set of genes, both dominant and recessive, possessed by a particular cell or organism.
genotyping—
The process of assessing genetic variations present in an individual.
hairpin—
Antiparallel duplex structure that forms by pairing of inverted repeat sequences within a single-stranded nucleic acid. The helical section is called the stem and the unpaired base segment at the end of the structure is called the loop.
hot-start pcr—
Technique that is commonly used to improve the sensitivity and specificity of PCR amplification. A hot start is performed by withholding from the reaction mix a key component necessary for amplification until the reaction reaches a temperature above the optimal annealing temperature of the primers. The component withheld from the reaction mix can be primers, DNA polymerase, MgCl2, or dNTPs.
hybridization—
The process of forming a double-stranded nucleic acid molecule, for example between a nucleotide sequence (probe) and a target.
ligation—
The process of joining two or more DNA fragments.
melting temperature (Tm)—
The temperature at which 50% of the DNA becomes single-stranded.
microarray—
Sets of miniaturized chemical reaction areas that are used to test DNA fragments, antibodies, or proteins. Usually the probes are immobilized on a chip and hybridized with target.
mismatch—
Unconventional base pairing (other than C with G, and A with T or U). A mismatched base pair has lower bonding energy and decreases the stability of the DNA molecule.
nucleic acid—
Linear polymers of nucleotides, linked by 3¢, 5¢ phosphodiester linkages. In DNA, deoxyribonucleic acid, the sugar group is deoxyribose, and the bases consist of adenine, guanine, thymine, and cytosine. RNA, ribonucleic acid, has ribose as the sugar, and uracil replaces thymine.
oligonucleotide—
Linear sequence comprising as many as 25 nucleotides joined by phosphodiester bonds, generally used as a DNA synthesis primer.
photobleaching—
Photobleaching is the irreversible destruction of a fluorophore in the excited state. Different fluorophores have different rates of photobleaching. For example, fluorescein photobleaches very easily. Often the rate of decomposition is proportional to the intensity of illumination. A simple and practical way to overcome this is to reduce the incident radiation.
polymerase—
An enzyme that catalyzes the synthesis of nucleic acids on pre-existing nucleic acid templates, assembling RNA from ribonucleotides or DNA from deoxyribonucleotides.
polymerase chain reaction (pcr)—
A laboratory technique that rapidly amplifies a specific region of double-stranded DNA, predetermined by the pair of primers used for amplification. Generally involves the use of a heat-stable DNA polymerase.
precision—
The degree of agreement among individual test results when the procedure is applied repeatedly to multiple samplings of a homogenous sample.
primer—
Nucleic acid polymerases link a mononucleotide to a chain of nucleic acids, which is called the primer. RNA polymerases are able to use a single nucleotide as a primer, but DNA polymerases always require an oligonucleotide.
probe—
A specific DNA or RNA sequence that has been labeled by radioactive, fluorescent, or chemiluminescent tags and is used to detect complementary sequences by hybridization techniques such as blotting or colony hybridization. In addition, probes can also be used for quantitation of amplicons as described for quantitative PCR assays. A more detailed description of such probes is given in the general information chapter, Nucleic Acid-Based Techniques—Amplification  1127
1127 .
.
processivity—
The ability of an enzyme to repetitively continue its catalytic function without dissociating from its substrate.
proofreading activity—
Literally to read for the purpose of detecting errors for later correction. DNA polymerase has a 3¢ to 5¢ exonuclease activity that is used during polymerization to remove recently added nucleotides that are incorrectly paired.
quantitation limit—
It is the lowest amount of analyte in a sample that can be determined with an acceptable precision and accuracy under the stated experimental conditions.
quenching—
The process of extinguishing, removing, or diminishing a physical property such as heat or light. Fluorescence quenching can be either collisional or static.
reverse transcriptase—
An enzyme that requires a DNA primer and catalyzes the synthesis of a DNA strand from an RNA template. An enzyme that can use RNA as a template to synthesize DNA.
reverse transcription (rt)—
The process of making cDNA using an RNA template.
real-time pcr—
May often be referred to as Quantitative PCR or Real-Time Quantitative PCR but not RT-PCR and is a procedure for simultaneous DNA quantitation and amplification. The generation of amplicons monitored as they are generated by the use of a fluorescent reporter system and captured by sophisticated instrumentation.
real-time (rt-pcr)—
The combination of real-time PCR and reverse transcription PCR.
reverse transcriptase polymerase chain reaction (rt-pcr)—
A variation of the PCR technique in which cDNA is made from RNA via reverse transcription. The cDNA is then amplified using standard PCR protocols.
ribonucleic acid (rna)—
A type of nucleic acid composed of a specific sequence of ribonucleotides linked together. RNA contains the following four ribonucleosides: A, C, G, and U.
robustness—
The robustness of an analytical procedure is a measure of its capacity to remain unaffected by small but deliberate variations in procedural parameters listed in the procedure documentation and provides an indication of its suitability during normal usage.
rTth dna polymerase—
Recombinant thermostable DNA polymerase originally isolated from the bacterium Thermus thermophilus. rTth has optimal activity at 70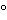 –80
–80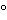 and survives the denaturation steps of PCR. In addition to DNA polymerase activity, it has efficient reverse transcriptase activity in the presence of manganese.
and survives the denaturation steps of PCR. In addition to DNA polymerase activity, it has efficient reverse transcriptase activity in the presence of manganese.
specificity—
The ability to assess unequivocally the analyte in the presence of components that may be expected to be present, such as impurities, degradation proeducts, and matrix components.
Taq dna—
Thermostable DNA polymerase that is originally isolated from the bacterium Thermus aquaticus, Taq has optimal activity at 70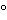 –80
–80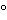 and is not degraded during the high-heat denaturation steps of PCR.
and is not degraded during the high-heat denaturation steps of PCR.
template—
A master copy used to start the DNA or RNA replication process.
transcription—
The synthesis of RNA using a DNA template.
ABBREVIATIONS
| AABB | American Association of Blood Banks |
| ACD | acid citrate dextrose |
| ASO | allele-specific oligonucleotides |
| bDNA | branched DNA assay |
| BMA | bone marrow aspirate |
| CE-LIF | capillary electrophoresis and laser-induced fluorescence |
| CCD | charge-coupled device |
| cDNA | complementary DNA |
| CPR | cyclic probe reaction |
| CsCl | cesium chloride |
| Ct | cycle threshold |
| DEPC | diethylpyrocarbonate |
| DHPLC | denaturing high-performance liquid chromatography |
| DNA | deoxyribonucleic acid |
| DNase | deoxyribonuclease |
| dNTP | deoxyribonucleotide triphosphate |
| DMSO | dimethyl sulfoxide |
| dNTP | dinucleotide triphosphate |
| DOP-PCR | degenerated oligonucleotide primed PCR |
| dsDNA | double-stranded DNA |
| ssDNA | single-stranded DNA |
| dUTP | 2¢-deoxyuridine 5¢-triphosphate |
| dTTP | 2¢-deoxythymidine 5¢-triphosphate |
| ESI | electrospray ionization |
| EDTA | ethylenediaminetetraacetic acid |
| FDA | Food and Drug Administration |
| FEN | flap endonuclease |
| FISH | fluorescent in situ hybridization |
| FFPE | formalin-fixed paraffin embedded |
| FRET | fluorescence resonance energy transfer |
| GLP | good laboratory practice |
| HCV | hepatitis C virus |
| HIV | human immunodeficiency virus |
| ICH | International Conference on Harmonization |
| LAPS | light-addressable potentiometric sensor |
| LCR | ligase chain reaction |
| LED | light-emitting diode |
| LNA | locked nucleic acid |
| MALDI | matrix-assisted laser desorption–ionization |
| MDA | multiple-displacement amplification |
| MOPS | 3-[N-morpholino]propanesulfonic acid |
| MS | mass spectrometry |
| mRNA | messenger RNA |
| NAT | nucleic acid amplification technologies |
| NASBA | nucleic acid sequence-based amplification |
| NTP | nucleotide triphosphate |
| OLA | oligonucleotide ligation assay |
| PAGE | polyacrylamide gel electrophoresis |
| PCR | polymerase chain reaction |
| PEP | primer-extension-preamplification |
| PPi | pyrophosphate |
| QA | quality assurance |
| QC | quality control |
| RCA | rolling circle amplification |
| RFLP | restriction fragment length polymorphism |
| RNA | ribonucleic acid |
| RNAse | ribonuclease |
| RT | reverse transcriptase |
| RT-PCR | reverse transcription-polymerase chain reaction |
| rTth | recombinant Thermus thermophilus |
| SDS | sodium dodecyl sulfate |
| SNP | single nucleotide polymorphism |
| 3SR | self-sustained sequence replication |
| SSCP | single-strand conformation polymorphism |
| STR | short tandem repeat |
| Taq | Thermus aquaticus |
| Tm | melting temperature; the temperature at which 50% of the double-stranded nucleic acid molecule becomes single-stranded |
| TMA | transcription-mediated amplification |
| TOF | time-of-flight |
| UNG | uracil-N-glycosylase |
| WGA | whole-genome amplification |
1
Reference materials for nucleic acid-based techniques are available from National Institute of Standards and Technology (NIST), http://ts.nist.gov/measurementservices/referencematerials/index.cfm.
Auxiliary Information— Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
| General Chapter | Anita Y. Szajek, Ph.D.
Senior Scientist 1-301-816-8325 |
(BBVV05) Biologics and Biotechnology - Vaccines and Virology |
USP32–NF27 Page 634
Pharmacopeial Forum: Volume No. 33(5) Page 984